Classifying T Cell Activity with Convolutional Neural Networks
Abstract
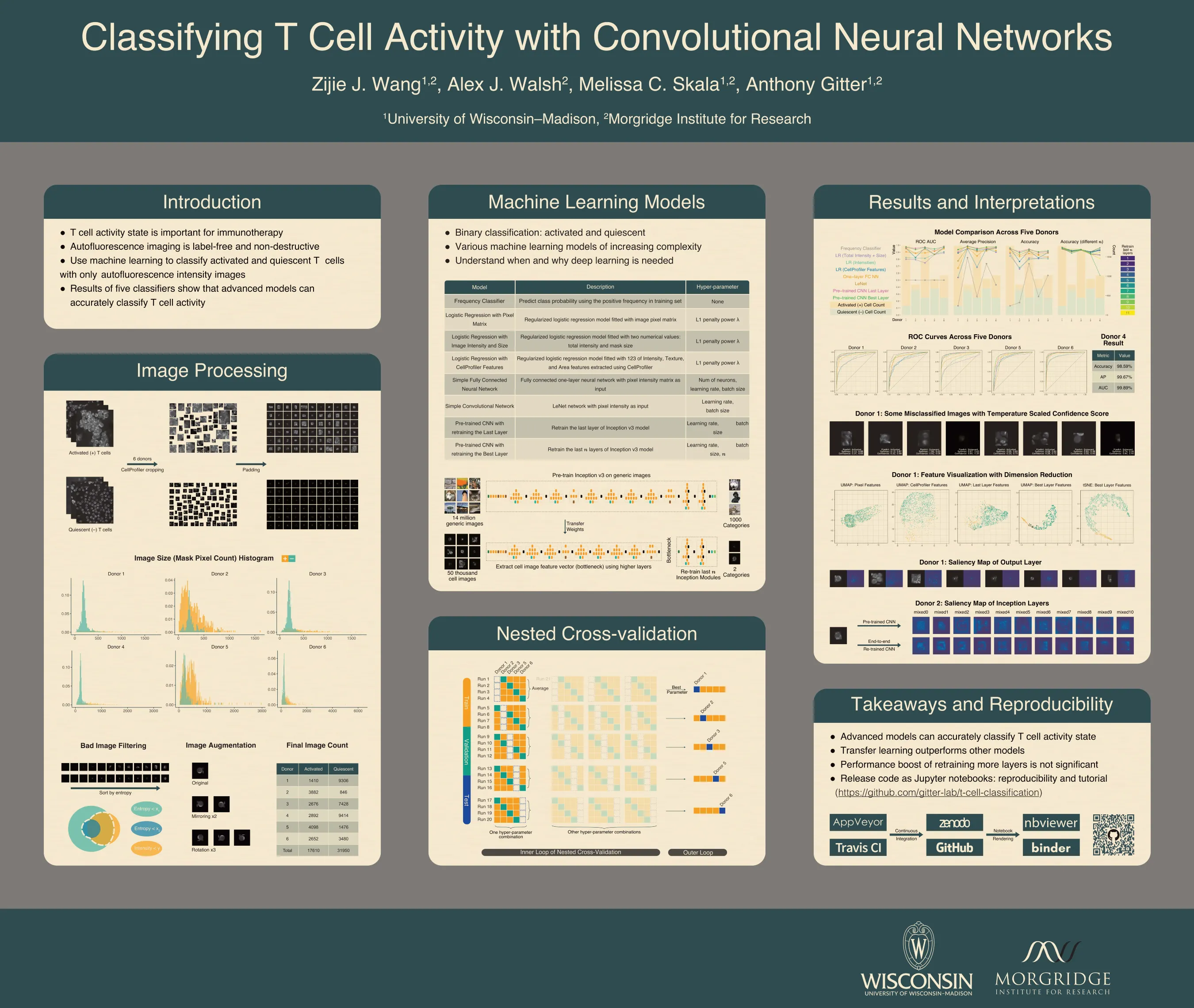
T cell activity state is an important component of immunotherapy efficacy in clinical cancer treatment. However, current image-based activity profiling methods destroy cells and require exogenous contrast agents, making them unsuitable for clinical applications. In this study, we use non-destructive, T cell autofluorescence microscopy images to measure NAD(P)H intensity and classify individual T cells as activated or quiescent. We assess five machine learning methods of increasing complexity, ranging from linear classifiers to deep convolutional neural networks pre-trained on generic images. To evaluate these models and determine whether they are accurate across different human T cell donors, we designed a meticulous nested cross-validation scheme to tune and test each model. A retrained convolutional neural network, the most advanced model, achieved an average accuracy of 91.4% when classifying quiescent and activated T cells. Importantly, it gave 98% accuracy on an independent donor that was held out until all aspects of the training and tuning procedures were finalized. This shows that autofluorescence microscopy with a state-of-the-art image classification algorithm is a powerful tool for label-free and non-destructive assessment of T cell activity state, even when only NAD(P)H intensity is provided as the input feature. In addition, our high-throughput hyperparameter selection results give empirical insights on practical deep learning deployment with microscopy image data. Similarly, the model comparisons examined the tradeoff between performance and model complexity, which provides alternative methods that are suitable when computing resource are limited. We observe that retraining more layers in a pre-trained convolutional neural network does not bring performance improvements that justify the high computational costs. Finally, we are preparing all of our code in Jupyter notebooks with reproducible examples of image processing and classification. These comprehensive notebooks serve as an instructional tool for readers who are not familiar with machine learning application on microscopy images.
Citation
Classifying T Cell Activity with Convolutional Neural Networks
@inproceedings{wang_classifying_poster_2019,
title = {Classifying {T} cell activity with convolutional neural networks},
language = {en},
conference = {International Society for Computational Biology Great Lakes Bioinformatics Conference},
author = {Wang, Zijie J. and Walsh, Alex J. and Skala, Melissa C. and Gitter, Anthony},
year = {2019}
}


